This will provide information like parameter names, covriates, etc from an rxode2 object.
This will provide information like parameter names, covriates, etc from an rxode2 object.
Value
List with the following elements.
isgood: Boolean variable indicating if the model is good.
msgs: Any messages from parsing the model.
elements: List with names of simulation elements:
covariates: Names of the covariates in the system.
parameters: Names of the parameters (subject level) in the system.
iiv: Names of the iiv parameters in the system.
states: Names of the states/compartments in the system.
txt_info: Summary information in text format.
list_info: Summary information in list format used with onbrand reporting.
ht_info: Summary information in HTML formot.
List with the following elements.
isgood: Boolean variable indicating if the model is good.
msgs: Any messages from parsing the model.
elements: List with names of simulation elements:
covariates: Names of the covariates in the system.
parameters: Names of the parameters (subject level) in the system.
iiv: Names of the iiv parameters in the system.
states: Names of the states/compartments in the system.
txt_info: Summary information in text format.
list_info: Summary information in list format used with onbrand reporting.
ht_info: Summary information in HTML formot.
Examples
# \donttest{
library(formods)
library(ggplot2)
#> Warning: package ‘ggplot2’ was built under R version 4.5.2
# For more information see the Clinical Trial Simulation vignette:
# https://ruminate.ubiquity.tools/articles/clinical_trial_simulation.html
# None of this will work if rxode2 isn't installed:
if(is_installed("rxode2")){
library(rxode2)
set.seed(8675309)
rxSetSeed(8675309)
my_model = function ()
{
description <- "One compartment PK model with linear clearance using differential equations"
ini({
lka <- 0.45
label("Absorption rate (Ka)")
lcl <- 1
label("Clearance (CL)")
lvc <- 3.45
label("Central volume of distribution (V)")
propSd <- c(0, 0.5)
label("Proportional residual error (fraction)")
etalcl ~ 0.1
})
model({
ka <- exp(lka)
cl <- exp(lcl + etalcl)
vc <- exp(lvc)
kel <- cl/vc
d/dt(depot) <- -ka * depot
d/dt(central) <- ka * depot - kel * central
Cc <- central/vc
Cc ~ prop(propSd)
})
}
# This creates an rxode2 object
object = rxode(my_model)
# If you want details about the parameters, states, etc
# in the model you can use this:
rxdetails = fetch_rxinfo(object)
rxdetails$elements
# Next we will create subjects. To do that we need to
# specify information about covariates:
nsub = 2
covs = list(
WT = list(type = "continuous",
sampling = "log-normal",
values = c(70, .15))
)
subs = mk_subjects(object = object,
nsub = nsub,
covs = covs)
head(subs$subjects)
rules = list(
dose = list(
condition = "TRUE",
action = list(
type = "dose",
state = "central",
values = "c(1)",
times = "c(0)",
durations = "c(0)")
)
)
# We evaulate the rules for dosing at time 0
eval_times = 0
# Stop 2 months after the last dose
output_times = seq(0, 56, 1)
# This runs the rule-based simulations
simres =
simulate_rules(
object = object,
subjects = subs[["subjects"]],
eval_times = eval_times,
output_times = output_times,
rules = rules)
# First subject data:
sub_1 = simres$simall[simres$simall$id == 1, ]
# First subjects events
evall = as.data.frame(simres$evall)
ev_sub_1 = evall[evall$id ==1, ]
# All of the simulation data
simall = simres$simall
simall$id = as.factor(simall$id)
# Timecourse
psim =
plot_sr_tc(
sro = simres,
dvcols = "Cc")
psim$fig
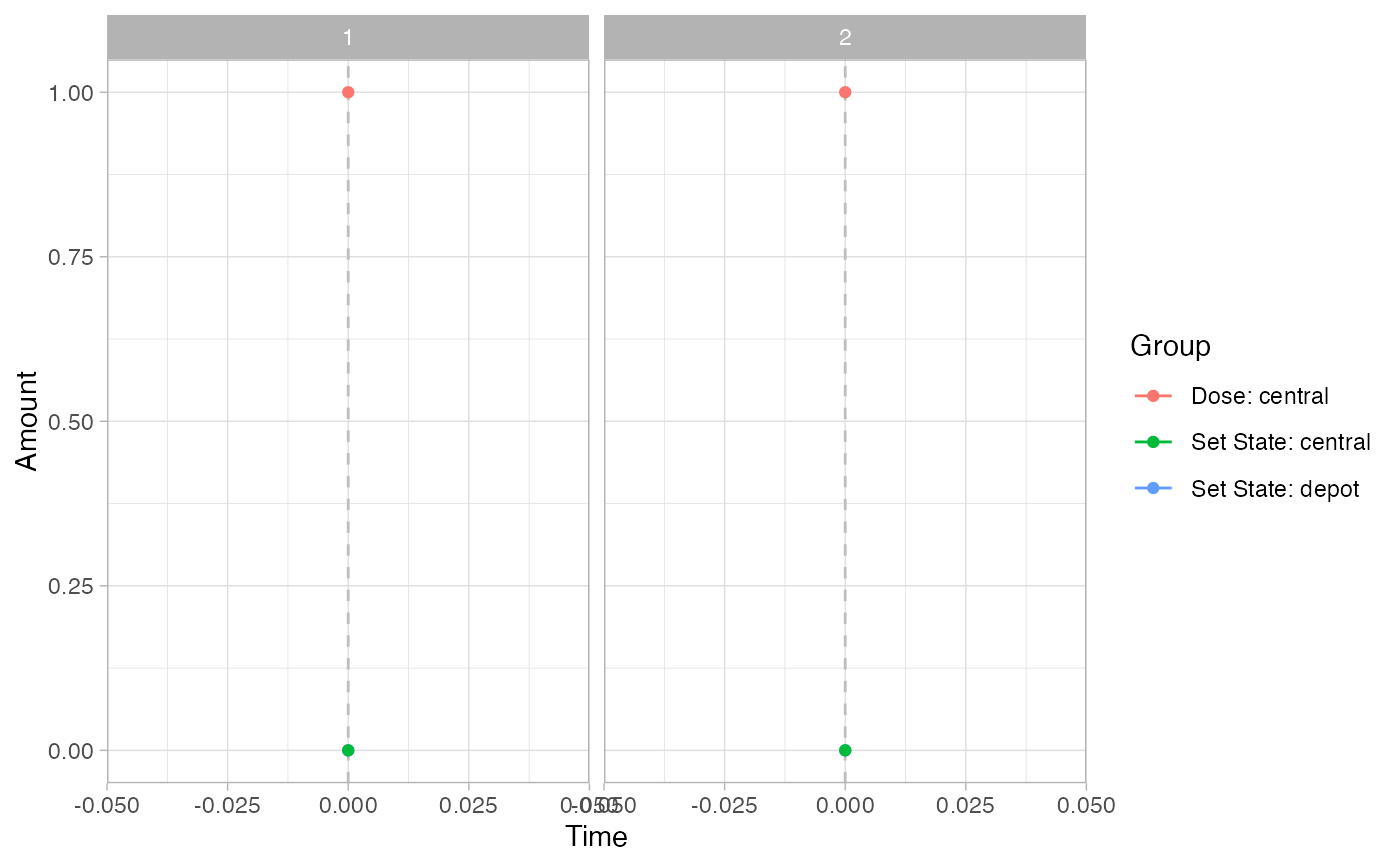
# Events
pev =
plot_sr_ev(
sro = simres,
ylog = FALSE)
pev$fig
}
#> rxode2 4.1.1 using 1 threads (see ?getRxThreads)
#> no cache: create with `rxCreateCache()`
#> ========================================
#> rxode2 has not detected OpenMP support and will run in single-threaded mode
#> This is a Mac. Please read https://mac.r-project.org/openmp/
#> ========================================
#>
#>
#> ℹ parameter labels from comments are typically ignored in non-interactive mode
#> ℹ Need to run with the source intact to parse comments
#>
#>
#> using C compiler: ‘Apple clang version 16.0.0 (clang-1600.0.26.6)’
#> using SDK: ‘’
#>
#>
#> Warning: multi-subject simulation without without 'omega'
#>
#>
#> using C compiler: ‘Apple clang version 16.0.0 (clang-1600.0.26.6)’
#> using SDK: ‘’
#> Warning: multi-subject simulation without without 'omega'
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?
 # }
# \donttest{
library(formods)
library(ggplot2)
# For more information see the Clinical Trial Simulation vignette:
# https://ruminate.ubiquity.tools/articles/clinical_trial_simulation.html
# None of this will work if rxode2 isn't installed:
if(is_installed("rxode2")){
library(rxode2)
set.seed(8675309)
rxSetSeed(8675309)
my_model = function ()
{
description <- "One compartment PK model with linear clearance using differential equations"
ini({
lka <- 0.45
label("Absorption rate (Ka)")
lcl <- 1
label("Clearance (CL)")
lvc <- 3.45
label("Central volume of distribution (V)")
propSd <- c(0, 0.5)
label("Proportional residual error (fraction)")
etalcl ~ 0.1
})
model({
ka <- exp(lka)
cl <- exp(lcl + etalcl)
vc <- exp(lvc)
kel <- cl/vc
d/dt(depot) <- -ka * depot
d/dt(central) <- ka * depot - kel * central
Cc <- central/vc
Cc ~ prop(propSd)
})
}
# This creates an rxode2 object
object = rxode(my_model)
# If you want details about the parameters, states, etc
# in the model you can use this:
rxdetails = fetch_rxinfo(object)
rxdetails$elements
# Next we will create subjects. To do that we need to
# specify information about covariates:
nsub = 2
covs = list(
WT = list(type = "continuous",
sampling = "log-normal",
values = c(70, .15))
)
subs = mk_subjects(object = object,
nsub = nsub,
covs = covs)
head(subs$subjects)
rules = list(
dose = list(
condition = "TRUE",
action = list(
type = "dose",
state = "central",
values = "c(1)",
times = "c(0)",
durations = "c(0)")
)
)
# We evaulate the rules for dosing at time 0
eval_times = 0
# Stop 2 months after the last dose
output_times = seq(0, 56, 1)
# This runs the rule-based simulations
simres =
simulate_rules(
object = object,
subjects = subs[["subjects"]],
eval_times = eval_times,
output_times = output_times,
rules = rules)
# First subject data:
sub_1 = simres$simall[simres$simall$id == 1, ]
# First subjects events
evall = as.data.frame(simres$evall)
ev_sub_1 = evall[evall$id ==1, ]
# All of the simulation data
simall = simres$simall
simall$id = as.factor(simall$id)
# Timecourse
psim =
plot_sr_tc(
sro = simres,
dvcols = "Cc")
psim$fig
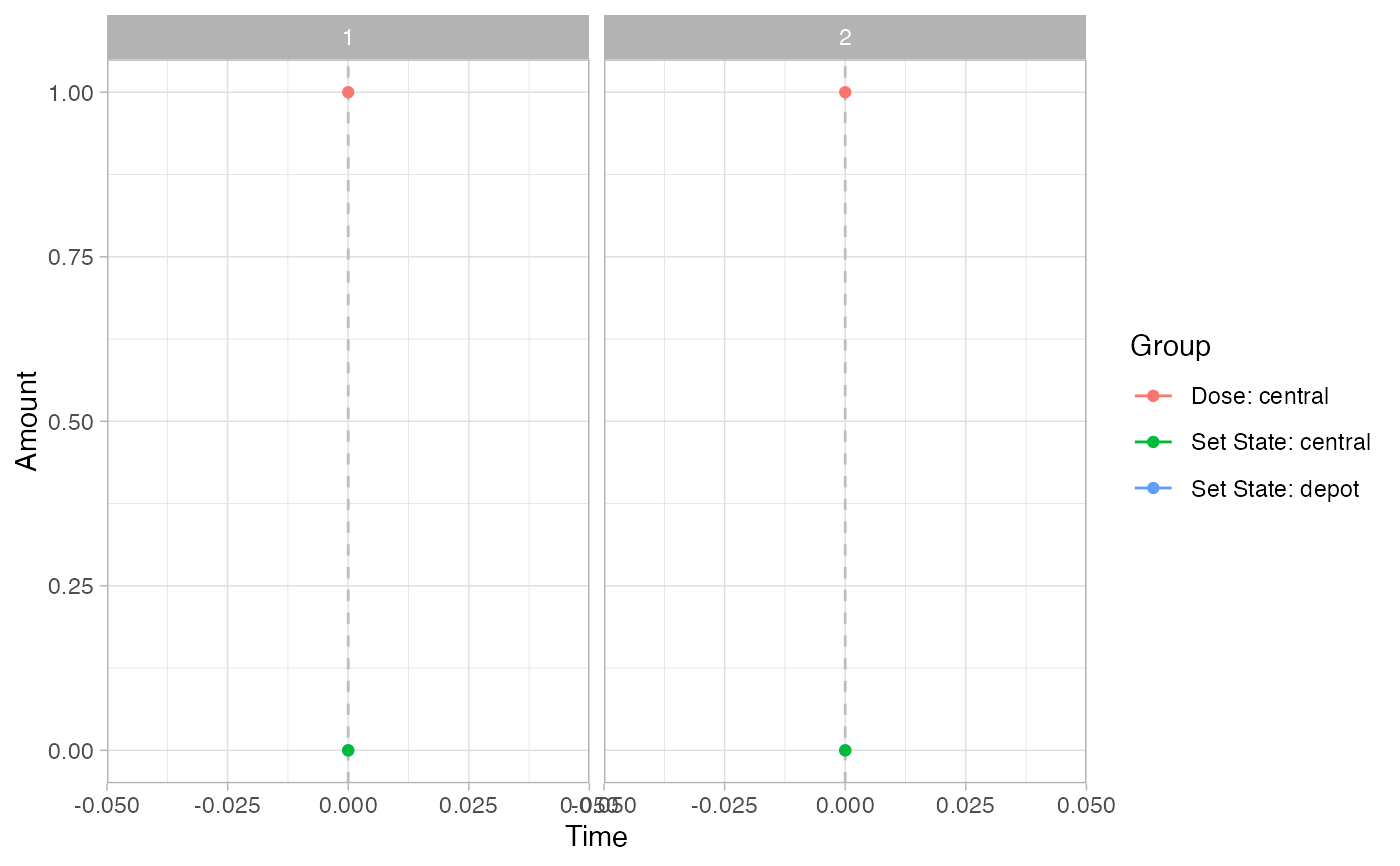
# Events
pev =
plot_sr_ev(
sro = simres,
ylog = FALSE)
pev$fig
}
#>
#>
#> ℹ parameter labels from comments are typically ignored in non-interactive mode
#> ℹ Need to run with the source intact to parse comments
#>
#>
#>
#>
#> Warning: multi-subject simulation without without 'omega'
#> Evaluation times ■■■■■■■■■■■■■■■■ 50% | ETA: 4s
#>
#>
#> Warning: multi-subject simulation without without 'omega'
#> Evaluation times ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ 100% | ETA: 0s
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?
# }
# \donttest{
library(formods)
library(ggplot2)
# For more information see the Clinical Trial Simulation vignette:
# https://ruminate.ubiquity.tools/articles/clinical_trial_simulation.html
# None of this will work if rxode2 isn't installed:
if(is_installed("rxode2")){
library(rxode2)
set.seed(8675309)
rxSetSeed(8675309)
my_model = function ()
{
description <- "One compartment PK model with linear clearance using differential equations"
ini({
lka <- 0.45
label("Absorption rate (Ka)")
lcl <- 1
label("Clearance (CL)")
lvc <- 3.45
label("Central volume of distribution (V)")
propSd <- c(0, 0.5)
label("Proportional residual error (fraction)")
etalcl ~ 0.1
})
model({
ka <- exp(lka)
cl <- exp(lcl + etalcl)
vc <- exp(lvc)
kel <- cl/vc
d/dt(depot) <- -ka * depot
d/dt(central) <- ka * depot - kel * central
Cc <- central/vc
Cc ~ prop(propSd)
})
}
# This creates an rxode2 object
object = rxode(my_model)
# If you want details about the parameters, states, etc
# in the model you can use this:
rxdetails = fetch_rxinfo(object)
rxdetails$elements
# Next we will create subjects. To do that we need to
# specify information about covariates:
nsub = 2
covs = list(
WT = list(type = "continuous",
sampling = "log-normal",
values = c(70, .15))
)
subs = mk_subjects(object = object,
nsub = nsub,
covs = covs)
head(subs$subjects)
rules = list(
dose = list(
condition = "TRUE",
action = list(
type = "dose",
state = "central",
values = "c(1)",
times = "c(0)",
durations = "c(0)")
)
)
# We evaulate the rules for dosing at time 0
eval_times = 0
# Stop 2 months after the last dose
output_times = seq(0, 56, 1)
# This runs the rule-based simulations
simres =
simulate_rules(
object = object,
subjects = subs[["subjects"]],
eval_times = eval_times,
output_times = output_times,
rules = rules)
# First subject data:
sub_1 = simres$simall[simres$simall$id == 1, ]
# First subjects events
evall = as.data.frame(simres$evall)
ev_sub_1 = evall[evall$id ==1, ]
# All of the simulation data
simall = simres$simall
simall$id = as.factor(simall$id)
# Timecourse
psim =
plot_sr_tc(
sro = simres,
dvcols = "Cc")
psim$fig
# Events
pev =
plot_sr_ev(
sro = simres,
ylog = FALSE)
pev$fig
}
#>
#>
#> ℹ parameter labels from comments are typically ignored in non-interactive mode
#> ℹ Need to run with the source intact to parse comments
#>
#>
#>
#>
#> Warning: multi-subject simulation without without 'omega'
#> Evaluation times ■■■■■■■■■■■■■■■■ 50% | ETA: 4s
#>
#>
#> Warning: multi-subject simulation without without 'omega'
#> Evaluation times ■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■■ 100% | ETA: 0s
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?
 # }
# }
